Branch evolution¶
Notebook contributors: William Albert, Hannah Weiser, July 2025
This notebook demonstrates the preparation of the 4DGeo branch evolution example. The dataset contains weekly LiDAR scans of a tree over a time period of 6 weeks, with the first scan on 27 March 2025 and the last scan on 9 May 2025.
The starting point of the notebook are pointwise motion vectors computed for all pairs of consecutive scans. These may be computed using tools like PlantMove. Here, we read these motion vectors from a .npz file with Python. In addition, we have .laz files for the individual scans, which for each epoch are split into the branch of interest and the rest of the tree.
Related publications:
- Di Wang, Puttonen, E., & Casella, E. (2022). PlantMove: A tool for quantifying motion fields of plant movements from point cloud time series. International Journal of Applied Earth Observation and Geoinformation, 110, 102781. https://doi.org/10.1016/j.jag.2022.102781
- Albert, W., Weiser, H., Tabernig, R., & Höfle, B. (2025). Wind during terrestrial laser scanning of trees: Simulation-based assessment of effects on point cloud features and leaf-wood classification. ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences, X-G-2025, 25–32. https://doi.org/10.5194/isprs-annals-x-g-2025-25-2025
- Weiser, H., & Höfle, B. (2025). Advancing vegetation monitoring with virtual laser scanning of dynamic scenes (VLS-4D): Opportunities, implementations and future perspectives. https://doi.org/10.31223/x51q5v
Imports¶
import os
import sys
import json
import numpy as np
import matplotlib.pyplot as plt
from scipy.spatial import KDTree
import pandas as pd
import laspy
import vapc
from vapc.las_split_append_merge import las_merge
sys.path.insert(0, "../src")
from fourdgeo import projection
from fourdgeo import utilities
# File download and handling
from pathlib import Path
import pooch
from tqdm import tqdm
# Image and geometry handling
from PIL import Image, ImageDraw, ImageFont, ImageSequence
from IPython.display import HTML
from shapely.geometry import shape
Get the data¶
# Handle file download/reading here
data_url = "https://zenodo.org/api/records/16153369/files/branch_evolution.zip/content"
data_hash = "645670e7f71227df47cf1a41a281aaf052a299fc5462a048503d33b6db1009be"
file_name = "branch_evolution.zip"
data_folder = "data/branch_evolution"
if not Path(data_folder).exists():
fnames = pooch.retrieve(url=data_url,
known_hash=data_hash,
path="./",
fname=file_name,
processor=pooch.Unzip(extract_dir=data_folder),
progressbar=True)
os.remove(file_name)
Overview of the data¶
Let's first get an overview of the input data. We visualize the point clouds of the first and last scan side by side with matplotlib by height, showing the branch of interest in black.
laz_files_branches = list(Path(data_folder).glob("*_branch.laz"))
laz_files_rest = list(Path(data_folder).glob("*_rest.laz"))
las_b0 = laspy.read(laz_files_branches[0])
pc_b0 = np.c_[las_b0.x, las_b0.y, las_b0.z]
las_b6 = laspy.read(laz_files_branches[6])
pc_b6 = np.c_[las_b6.x, las_b6.y, las_b6.z]
las_r0 = laspy.read(laz_files_rest[0])
pc_r0 = np.c_[las_r0.x, las_r0.y, las_r0.z]
las_r6 = laspy.read(laz_files_rest[6])
pc_r6 = np.c_[las_r6.x, las_r6.y, las_r6.z]
fig = plt.figure(figsize=(16,8))
# Scatter plot
ax = fig.add_subplot(121, projection="3d")
sc1 = ax.scatter(
pc_b0[::5, 0],
pc_b0[::5, 1],
pc_b0[::5, 2],
color="black", s=0.02
)
sc1_2 = ax.scatter(
pc_r0[::5, 0],
pc_r0[::5, 1],
pc_r0[::5, 2],
c=pc_r0[::5, 2], s=0.02
)
ax2 = fig.add_subplot(122, projection="3d")
sc2 = ax2.scatter(
pc_b6[::5, 0],
pc_b6[::5, 1],
pc_b6[::5, 2],
color="black", s=0.02
)
sc2_2 = ax2.scatter(
pc_r6[::5, 0],
pc_r6[::5, 1],
pc_r6[::5, 2],
c=pc_r6[::5, 2], s=0.02
)
# Add axis labels.
ax.set_zlabel("[m]")
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.set_title("2025-03-27")
ax2.set_zlabel("[m]")
ax2.set_xticklabels([])
ax2.set_yticklabels([])
ax2.set_title("2025-05-09")
# set equal axes
box = (np.ptp(pc_r6[:, 0]), np.ptp(pc_r6[:, 1]), np.ptp(pc_r6[:, 2]))
ax.set_box_aspect(box)
ax2.set_box_aspect(box)
# Set title.
# Set view
ax.view_init(elev=5, azim=10, roll=0)
ax2.view_init(elev=5, azim=10, roll=0)
# Display results
fig.suptitle("Comparison of first and last epoch", fontsize=18)
plt.show()
The motion vectors have been computed with PlantMove for each point in a selected branch in the point cloud. They are stored in a .npz archive. Let's look at the structure for the first observation (change between first epoch on 27 March and second epoch on 4 April 2025).
cloud_x is the first time step,
cloud_y is the second time step,
motionfield is the derived motion field
and cloud_new is the point cloud of the second time step registered onto the first time step by applying the motion vectors.
files = list(Path(data_folder).glob("*.npz"))
files = sorted(files)
obs_file = files[0]
data = np.load(obs_file)
cloud_x = data["x_original"] # t0
cloud_y = data["y_original"] # t1
cloud_new = data["y_new"] # t1 to t0
motionfield = data["motionfield"] # motion field from t0 to t1
motion_vec = motionfield[:, 3:6]
motion_magnitudes = np.linalg.norm(motion_vec, axis=1)
print("Motion magnitudes between t0 and t1:\n")
print(f"{'Min:':20} {motion_magnitudes.min():.3f} m")
print(f"{'Max:':20} {motion_magnitudes.max():.3f} m")
print(f"{'Mean:':20} {motion_magnitudes.mean():.3f} m")
print(f"{'Standard deviation:':20} {motion_magnitudes.std():.3f} m")
Motion magnitudes between t0 and t1: Min: 0.000 m Max: 0.018 m Mean: 0.005 m Standard deviation: 0.003 m
Projections (point cloud)¶
For each observation, we create a 2D image of the point cloud by projecting the 3D points onto a 2D plane from the viewpoint of the scanner. Before that, we need to prepare the data accordingly.
Prepare the configuration file¶
We use a configuration dictionary that contains general project settings like the output_folder and the relevant settings for the point cloud projection. For the projection, the camera_position is essential, for which we choose the location of the scanner (in ETRS89, UTM zone 32 N).
configuration = {
"project_setting": {
"project_name": "Branch_evolution",
"output_folder": "./out/branch_evolution",
"temporal_format": "%y%m%d_%H%M%S",
"silent_mode": True,
"include_timestamp": False
},
"pc_projection": {
"pc_path": "",
"make_range_image": False,
"make_color_image": True,
"top_view": False,
"save_rot_pc": False,
"resolution_cm": 0.2,
"camera_position": [
475923.4099,
5473923.7662,
159.1088
],
"rgb_light_intensity": 100,
"range_light_intensity": 60,
"epsg": 25832
}
}
Merge each branch with its respective tree¶
In our input data, point clouds of the branch of interest are separated from the rest of the tree. For each epoch, we merge
- The branch point cloud of the epoch (coloured in red),
- The branch point cloud of the next epoch (coloured in blue)
- The point cloud of the remaining tree at the epoch (coloured in grayscale by intensity)
laz_files_rest = sorted(laz_files_rest)
laz_files_branches = sorted(laz_files_branches)
pcs = []
for enum in np.arange(len(laz_files_rest)-1):
pos = str(laz_files_rest[enum]).find("_icp_rest")
new_file_path = str(laz_files_rest[enum])[:pos]
with laspy.open(laz_files_branches[enum]) as las_branch_A:
las_branch_A = las_branch_A.read()
las_branch_A.red = np.full(len(las_branch_A.points), 65535)
las_branch_A.green = np.full(len(las_branch_A.points), 22768)
las_branch_A.blue = np.full(len(las_branch_A.points), 0)
las_branch_A.write(laz_files_branches[enum])
with laspy.open(laz_files_branches[enum+1]) as las_branch_B:
las_branch_B = las_branch_B.read()
las_branch_B.red = np.full(len(las_branch_B.points), 0)
las_branch_B.green = np.full(len(las_branch_B.points), 0)
las_branch_B.blue = np.full(len(las_branch_B.points), 65535)
las_branch_B.write(laz_files_branches[enum+1])
with laspy.open(laz_files_rest[enum]) as las_rest_A:
las_rest_A = las_rest_A.read()
intensities = las_rest_A.intensity
# Normalize intensities to 0-65535
norm = (intensities - intensities.min()) / (np.ptp(intensities) + 1e-8)
colors = (norm * 65535).astype(np.uint16)
# Assign grayscale color based on intensity
las_rest_A.red = las_rest_A.green = las_rest_A.blue = colors
las_rest_A.write(laz_files_rest[enum])
las_merge([laz_files_rest[enum], laz_files_branches[enum], laz_files_branches[enum+1]], f"{new_file_path}_{enum}_{enum+1}.laz", point_source_id=True)
pcs.append(f"{new_file_path}_{enum}_{enum+1}.laz")
Project the point clouds onto 2D images¶
We now generate the background images. For this, we are using classes and functions from the fourdgeo library. The class PCloudProjection directly takes our configuration file as input and writes the generated images to our specific output_folder.
images = []
list_background_projections = []
for enum, pc in enumerate(pcs):
lf = laspy.read(pc)
configuration['pc_projection']['pc_path'] = pc
project_name = configuration['project_setting']['project_name']
output_folder = configuration['project_setting']['output_folder']
background_projection = projection.PCloudProjection(
configuration = configuration,
project_name = project_name,
projected_image_folder = output_folder
)
# First projection
if enum == 0:
(
ref_h_fov, ref_v_fov, ref_anchor_point_xyz,
ref_h_img_res, ref_v_img_res
) = background_projection.project_pc(buffer_m = 0.5)
# Next projections using reference data
else:
background_projection.project_pc(
ref_theta=ref_h_fov[0],
ref_phi=ref_v_fov[0],
ref_anchor_point_xyz=ref_anchor_point_xyz,
ref_h_fov=ref_h_fov,
ref_v_fov=ref_v_fov,
ref_h_img_res=ref_h_img_res,
ref_v_img_res=ref_v_img_res,
buffer_m = 0.5
)
bg_img = background_projection.bg_image_filename[0]
if bg_img[0] == ".":
bg_img = bg_img[2:]
outfile = bg_img.split('.')[0] + f"_{enum}_{enum+1}." + bg_img.split('.')[1]
try:
os.rename(bg_img, outfile)
except FileExistsError:
os.remove(outfile)
os.rename(bg_img, outfile)
images.append(outfile)
background_projection.bg_image_filename[0] = outfile
list_background_projections.append(background_projection)
Convert .tif Images to .png Images¶
Because the dashboard (currently) does not support TIF files, we need to convert the generated background images to the PNG format.
png_images = []
for background_projection in list_background_projections:
image_path = background_projection.bg_image_filename[0]
filename = str.split(image_path, ".")[0]
try:
with Image.open(image_path) as im:
for i, page in enumerate(ImageSequence.Iterator(im)):
out_path = filename + ".png"
if not os.path.isfile(out_path):
try:
page.save(out_path)
except:
print(out_path)
png_images.append(out_path)
except:
print(filename)
Let's create a GIF from the images and display it.
gif_path = "../docs/img/tree_projections.gif"
frames = [Image.open(img).convert("RGB") for img in images]
frames[0].save(
gif_path,
save_all=True,
append_images=frames[1:],
duration=500,
loop=0
)
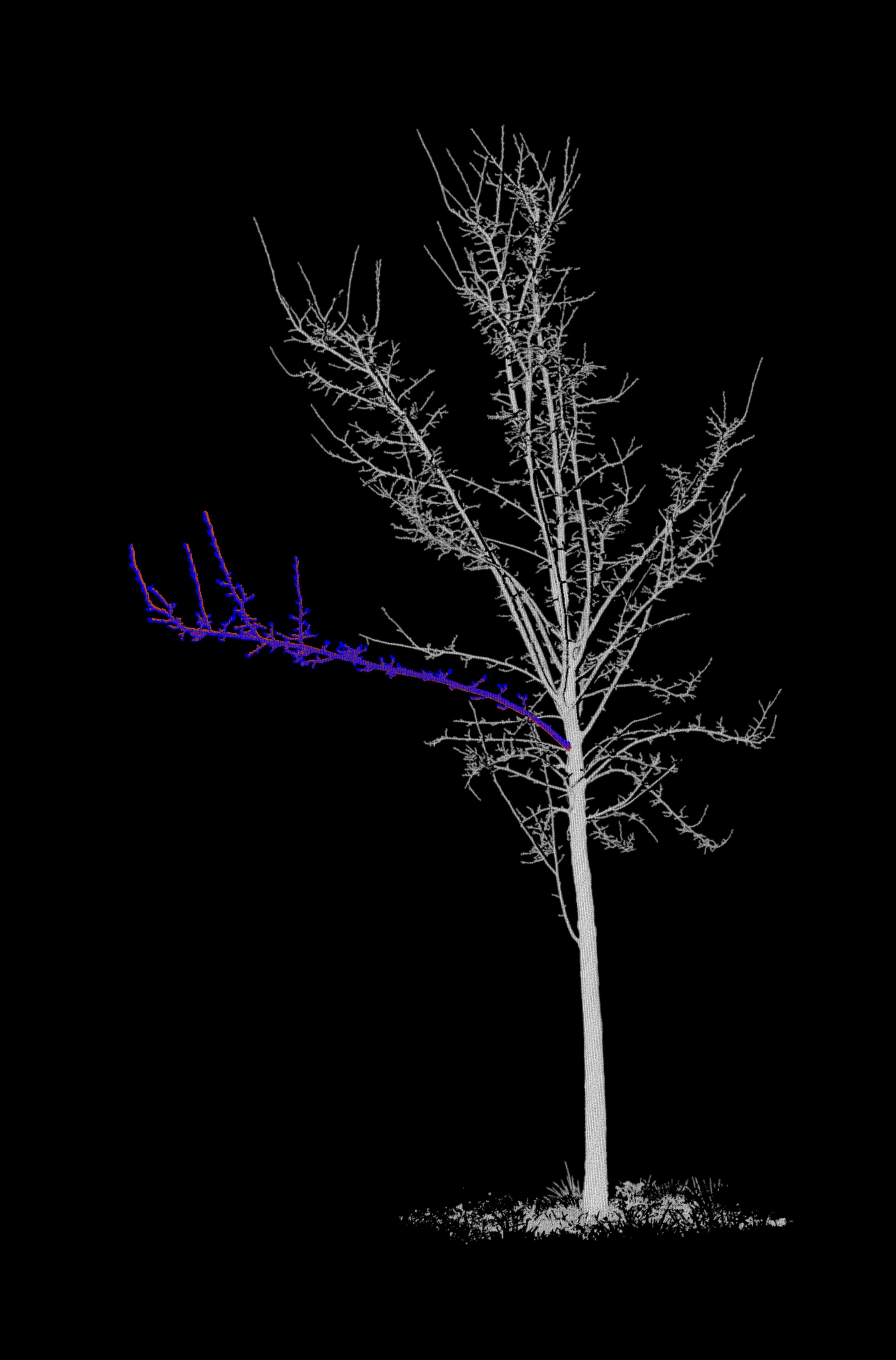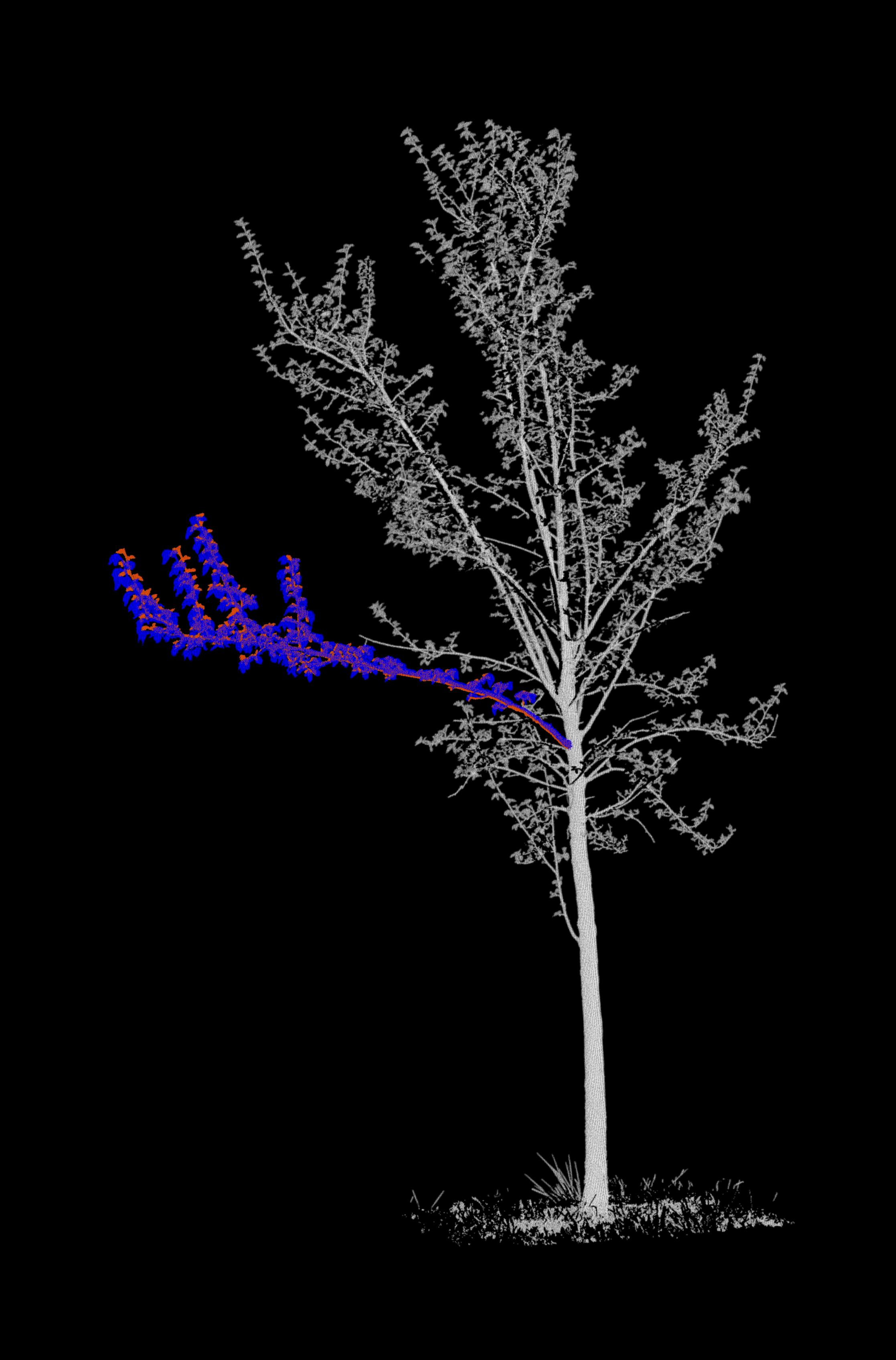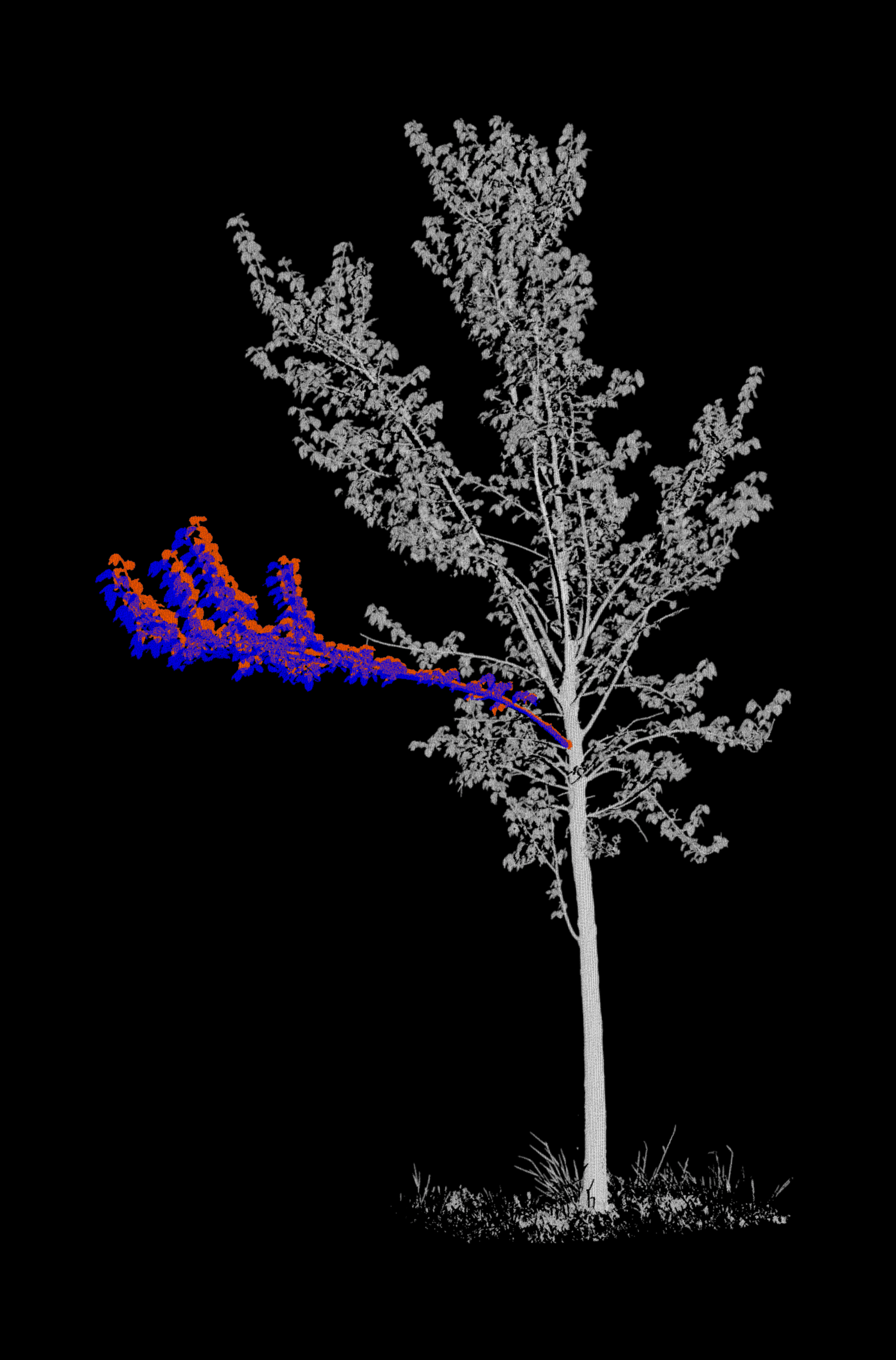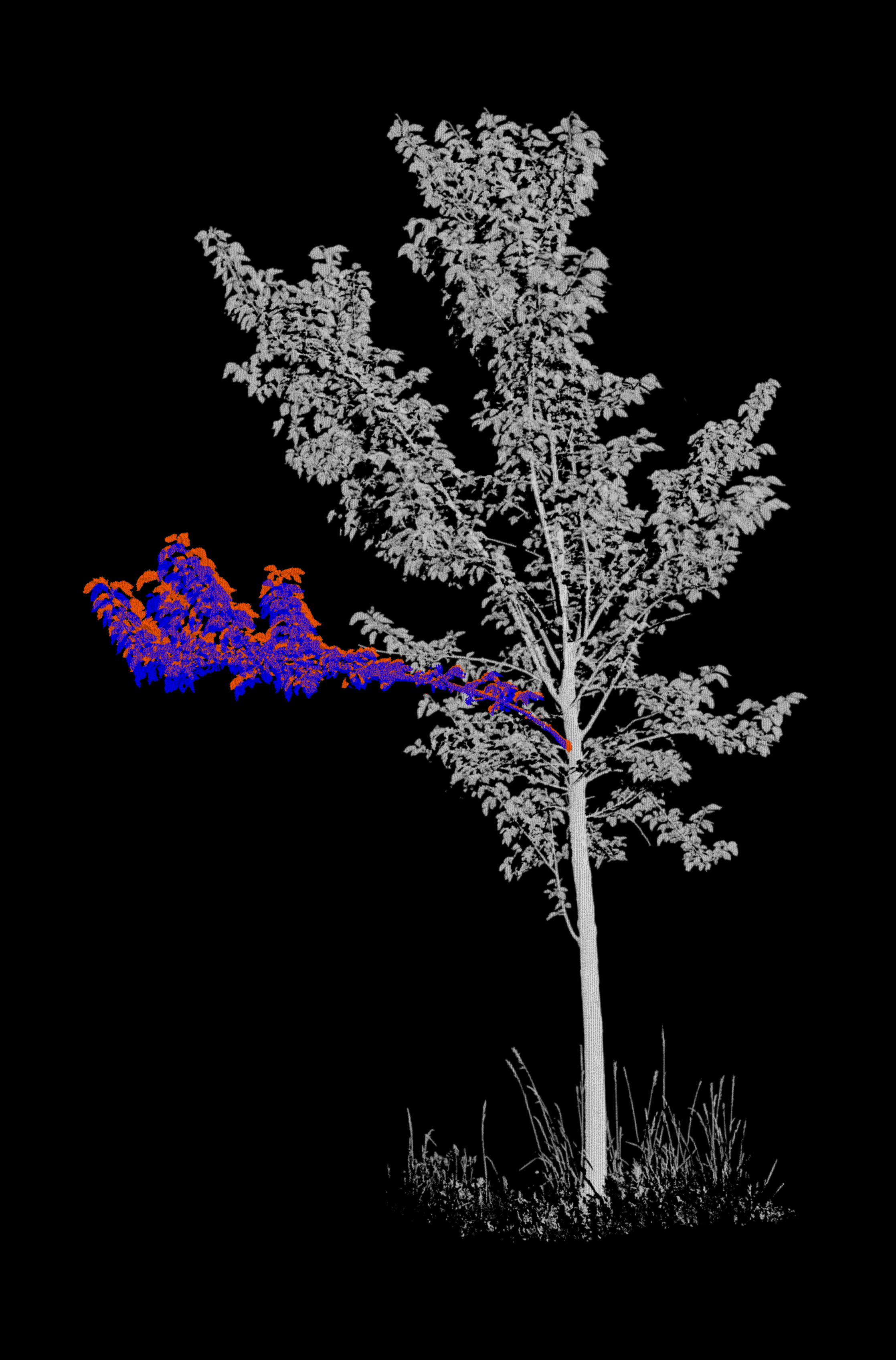
Projections (motion vectors)¶
Derive a subset of motion vectors by voxelization and save as geoObject¶
Since the visualization would be too cluttered with all motion vectors, we reduce the number of motion vectors by voxelizing the point cloud and only keeping one motion vector per voxel. We write this motion vector as a dictionary into a list observations.
if configuration["project_setting"]["silent_mode"]:
vapc.enable_trace(False)
vapc.enable_timeit(False)
voxel_size = 0.05
project_name = configuration["project_setting"]["project_name"]
observations = {"observations": []}
for epoch_id, file in enumerate(files):
geoObjects = []
# extract time from filenames
t_min = utilities.iso_timestamp("_".join(file.stem.split("_")[1:3]))
t_max = utilities.iso_timestamp("_".join(file.stem.split("_")[7:9]))
data = np.load(file)
cloud_x = data["x_original"] # t0
cloud_y = data["y_original"] # t1
cloud_new = data["y_new"] # t1 to t0
motionfield = data["motionfield"]
motion_vec = motionfield[:, 3:6]
motion_magnitudes = np.linalg.norm(motion_vec, axis=1)
if epoch_id == 0:
min_point = np.min(cloud_x, axis=0)
local_origin = np.floor(min_point / 100) * 100
# array to dataframe
df_y = pd.DataFrame(cloud_y, columns=["X", "Y", "Z"])
df_new = pd.DataFrame(cloud_new, columns=["X", "Y", "Z"])
vapc_y = vapc.Vapc(voxel_size=voxel_size, origin=local_origin.tolist())
vapc_y.df = df_y
vapc_y.compute = ["center_of_voxel"]
vapc_y.compute_requested_attributes()
vapc_new = vapc.Vapc(voxel_size=voxel_size, origin=local_origin.tolist())
vapc_new.df = df_new
vapc_new.compute = ["center_of_voxel"]
vapc_new.compute_requested_attributes()
if epoch_id == 0:
seed_points = np.array((vapc_new.df["center_x"].values, vapc_new.df["center_y"].values, vapc_new.df["center_z"].values)).T
seed_points = np.unique(seed_points, axis=0)
else:
seed_points = pts_y_original
# get nearest neighbours to seed points in cloud_new
tree_ynew = KDTree(cloud_new)
dist, idx_yn = tree_ynew.query(seed_points, k=1)
# get points
pts_y_original = cloud_y[idx_yn, :]
pts_y_new = cloud_new[idx_yn, :]
# create lines from nearest points to seed in cloud y and corresponding points in cloud x
lines = np.hstack((pts_y_original, pts_y_new))
# create json object for each line
for i in range(lines.shape[0]):
geoObject = {}
geoObject["id"] = f"{epoch_id}{i:04d}"
geoObject["type"] = "motion_vector"
geoObject["dateTime"] = t_min
geoObject["geometry"] = {
"type": "",
"coordinates": [
lines[i, 0:3].tolist(),
lines[i, 3:6].tolist()
]
}
geoObject["customAttributes"] = {
"motion_magnitudes": motion_magnitudes[i],
"epoch_id": epoch_id
}
geoObjects.append(geoObject)
observations["observations"].append({
"startDateTime": t_min,
"endDateTime": t_max,
"geoObjects": geoObjects,
"backgroundImageData": {
"url": list_background_projections[epoch_id].bg_image_filename[0],
"height": Image.open(images[epoch_id]).convert("RGB").size[1],
"width": Image.open(images[epoch_id]).convert("RGB").size[0]
},
})
Project the vectors from each observation onto their respective background image¶
Finally, we project these motion vectors onto the 2D image plane and save them in GeoJSON files in the output_folder, together with the projected images.
list_observation_projection = []
for epoch_id, observation in enumerate(observations['observations']):
background_projection = list_background_projections[epoch_id]
observation_projection = projection.ProjectChange(observation=observation,
project_name=f"{project_name}_{epoch_id}_{epoch_id+1}",
projected_image_path=background_projection.bg_image_filename[0],
projected_events_folder=output_folder,
epsg=None)
observation_projection.project_change()
list_observation_projection.append(observation_projection)
Let's display the branch (projected image) and the motion vectors in another GIF.
branch_crop_box = (130, 1170, 1600, 2000) # (left, upper, right, lower)
frames = []
gif_path = "../docs/img/tree_projections_plus_observations.gif"
font = ImageFont.load_default(size = 50)
for enum, img in enumerate(images):
frm = Image.open(img).convert("RGB")
frm = frm.crop(branch_crop_box)
draw = ImageDraw.Draw(frm)
draw.text((50, 50), f"Epoch: {enum}", fill=(255, 255, 255), font=font)
observation_projection = list_observation_projection[enum]
# Load geojson
with open(observation_projection.geojson_name, 'r') as f:
geojson_data = json.load(f)
for feature in geojson_data["features"]:
geom = shape(feature["geometry"])
coords = [(int(x) - branch_crop_box[0], -int(y) - branch_crop_box[1]) for x, y in geom.coords]
draw.line(coords, fill="yellow", width=2)
frames.append(frm)
frames[0].save(
gif_path,
save_all=True,
append_images=frames[1:],
duration=600,
loop=0
)
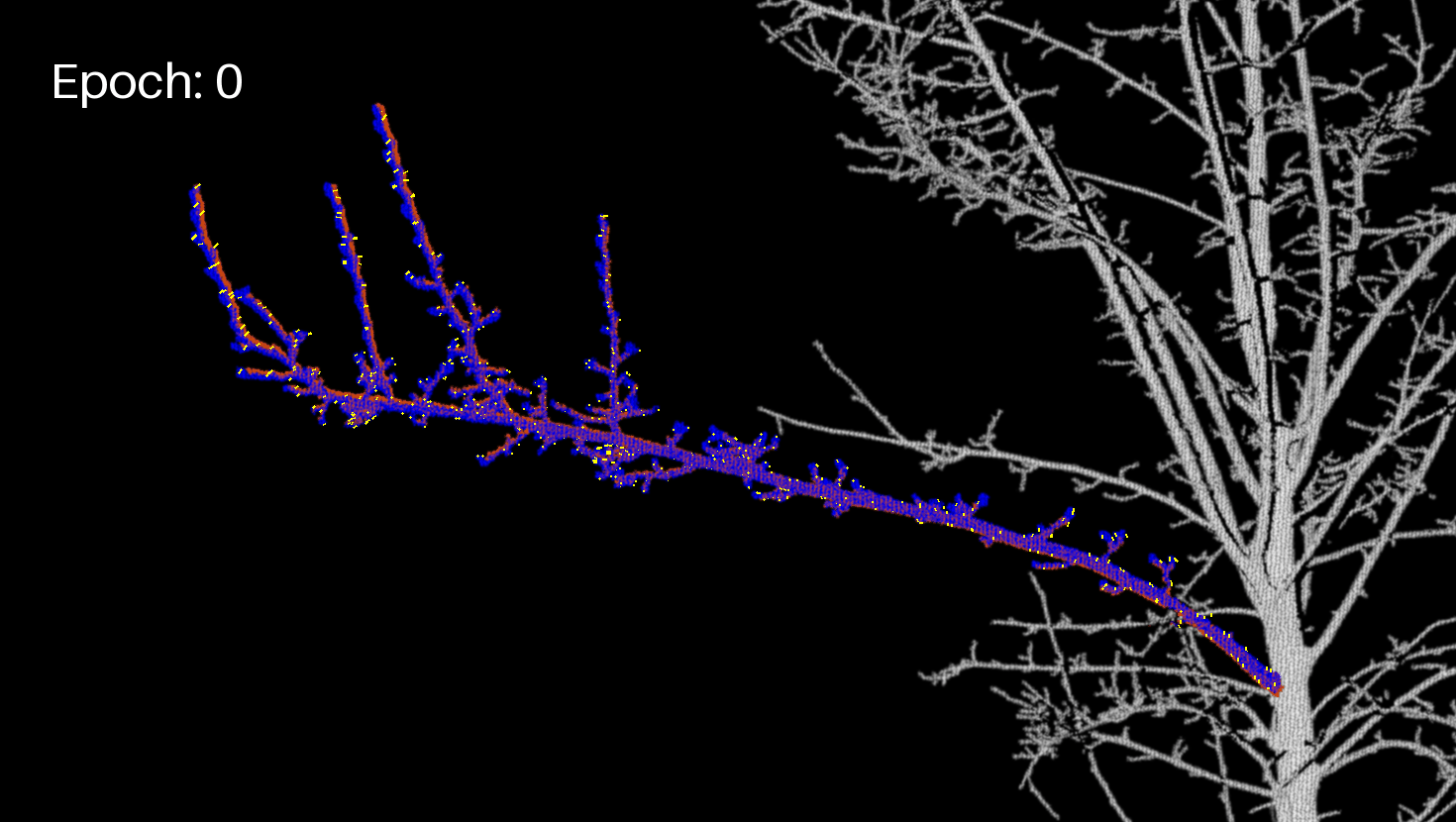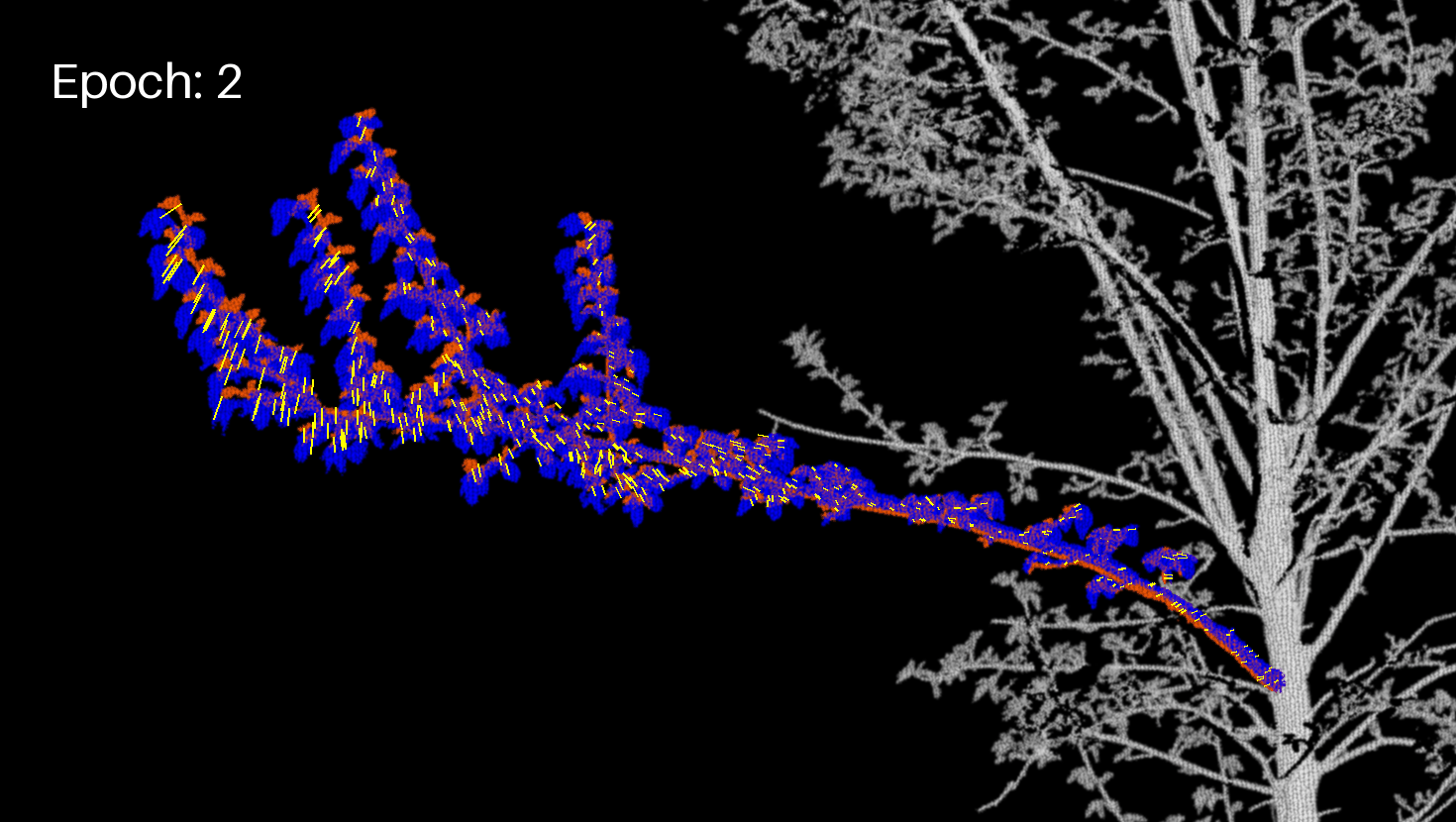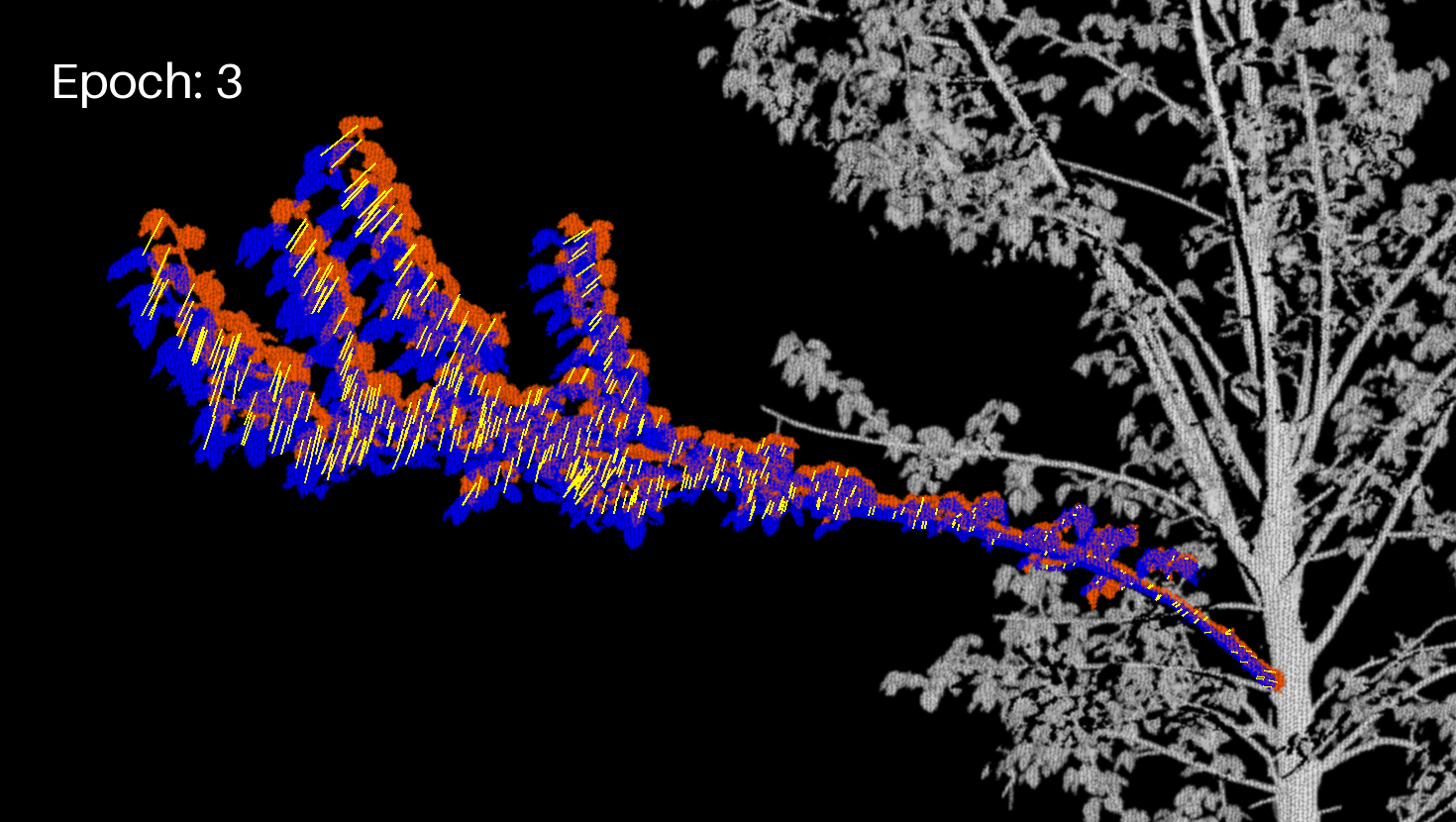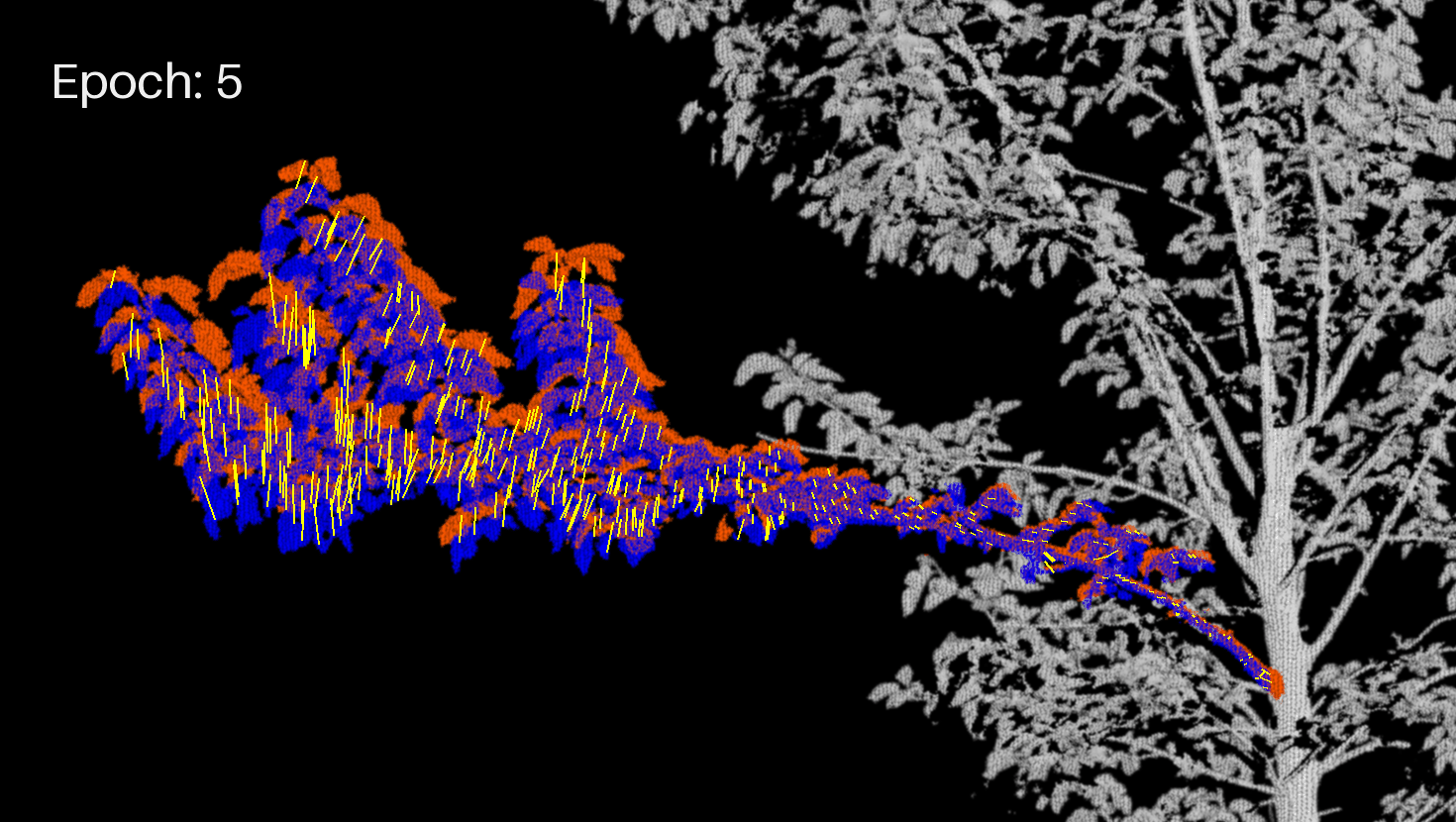
Extract the final JSON¶
In order to include this data into the dashboard, we now need to convert the created geojson data to the dashboard data model. For this, we iterate through all geojson files and create their observation objects and the aggregate them into one final data model object and store it in the output_folder. This files can then be loaded with the 4DGeo Dashboard.
aggregated_data = utilities.DataModel([])
for (i, observation_projection) in enumerate(list_observation_projection):
if observation_projection is None:
continue
elif observation_projection.observation["geoObjects"] is None:
img_size = Image.open(png_images[i]).convert("RGB").size
aggregated_data.observations.append(utilities.Observation(
startDateTime=observation_projection.observation["startDateTime"],
endDateTime=observation_projection.observation["endDateTime"],
geoObjects=[],
backgroundImageData=utilities.ImageData(
url=str("http://localhost:8001/" + png_images[i]).replace("\\", "/"),
width=img_size[0],
height=img_size[1]
)
))
continue
with open(observation_projection.geojson_name, 'r') as f:
geojson_data = json.load(f)
img_size = Image.open(png_images[i]).convert("RGB").size
geometry = geojson_data.get("features")[0].get("geometry")
coords = geometry.get("coordinates")
new_observations = utilities.convert_geojson_to_datamodel(
geojson=geojson_data,
bg_img=str("http://localhost:8001/" + png_images[i]).replace("\\", "/"),
width=img_size[0],
height=img_size[1]
)
aggregated_data.observations.extend(new_observations.observations)
with open(f"{output_folder}/final_data_model.json", "w") as f:
f.write(aggregated_data.toJSON())
Visualise the data in the dashboard¶
To see our results in the actual dashboard, we need to host the created json file to make it accessbile. As a quick and easy solution, we can use the http.server python library. The python script server-host.py hosts all the files in this directory. So in order to setup this local hosting, we need navigate to the 4DGeo/docs folder to execute the following command in a commandline:
python server_host.py
Lastly, inside of the dashboard, set the data source to http://localhost:8001/out/rockfall_monitoring/final_data_model.json.